SARA-Coffee Tutorial A quick start to SARA-Coffee structure based multiple RNA aligner.
Description
The web server allows the structure-based alignment of multiple RNAs combining the SARA [1] and the T-Coffee framework [2].
SARA executes a structural superposition of ribose C3' carbons to obtain the equivalences needed to return a structure-based alignment. As such, SARA acts as an aligner, therefore allowing its incorporation in a consistency-based environment like T-Coffee. The SARA-Coffee paper [3].
Data input
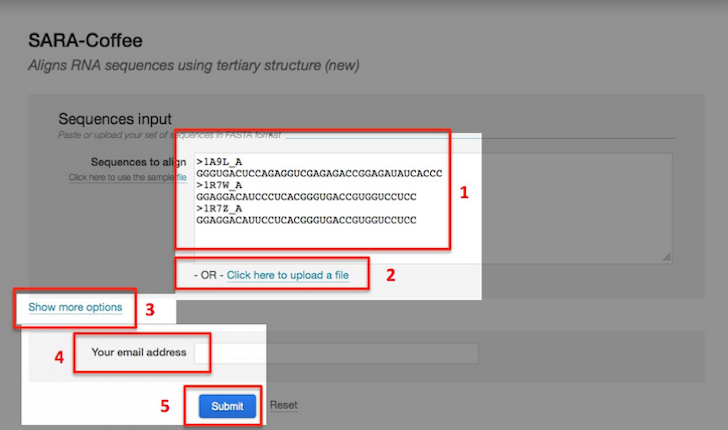
- Enter the sequences in the input field (marked with 1 in the picture below) or alternatively provide them using the Upload link (2).
-
The RNA sequences have to be in FASTA format. The sequence headers must indicate the structure PDB id
and the considered chain separated by an underscore (i.e. >PdbID_chain).
For instance the 5S ribosomal RNA available in the PDB structure 2J01 chain B should be passed to the server as:
>2J01_B UCCCCCGUGCCCAUAGCGGCGUGGAACCACCCGUUCCCAUUCCGAACACG GAAGUGAAACGCGCCAGCGCCGAUGGUACUGGGCGGGCGACCGCCUGGGA GAGUAGGUCGGUGCGGGGGAUU
- You may access to SARA-Coffee advanced settings by clicking the link Show more options (3).
- Provide your email address (4) if you want to be notified when the job complete.
- Press the button submit (5) to submit your request.
Please note that the computation can take some minutes to be fulfilled. You don't need to keep your browser open to wait for the result. You can close it or navigate away to another page.
Use the History link, in top-bar menu to get access to the computation result at any moment.
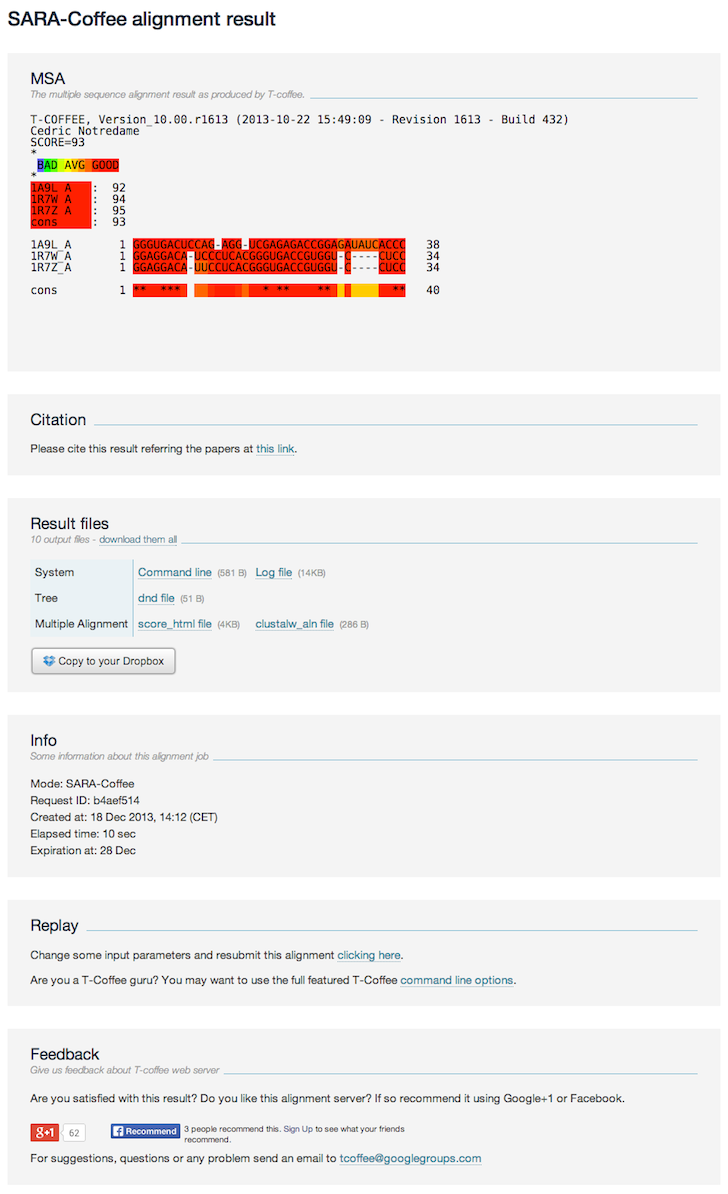
Output interpretation
The result page contains the following sections:
- MSA The MSA is colored according to the SARA-Coffee scheme (it is not a T-Coffee consistency based color scheme). Red bits are very reliable, while green and blue bits are unreliable. The details of the coloring scheme are given in a book chapter [4]
- Result files The result files coming from the alignment and Sara-Coffee method; all files can be downloaded as a single zip file
- Citation The related article when citing the SARA-Coffee.
- Info Some information related to the submitted job (running time, date, etc).
- Replay this link allows the user to re-run the job while modifying input options or data.
- Feedback To share your alignment experience in the social network.
Data input
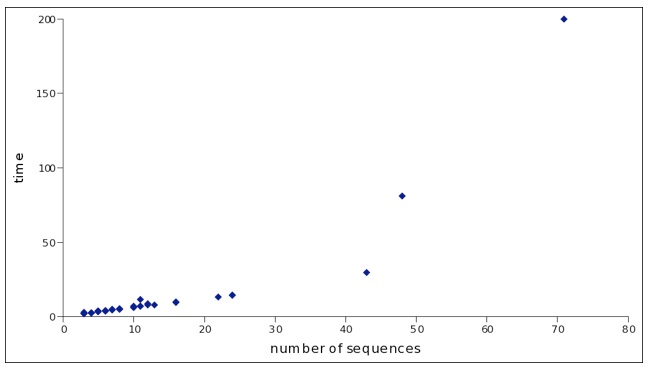
Running SARA-Coffee can be time expensive. SARA-Coffee web-server does not apply limitations in terms of number of input sequences and length but be aware that the computation can take long for larger RNA sets.
As a reference, see the figure below taken from SARA-Coffee paper. The x-axis shows the number of sequences included in each dataset. The vertical axis represents the single processor running time (user time plus sys time) expressed in seconds. The blue dots the RNA clusters analyzed.
References
-
RNA structure alignment by a unit-vector approach
Capriotti E, Marti-Renom MA -
T-Coffee: A novel method for fast and accurate multiple sequence alignment
Notredame C, Higgins DG, Heringa J -
Using tertiary structure for the computation of highly accurate multiple RNA alignments with the SARA-Coffee package
Kemena C, Bussotti G, Capriotti E, Marti-Renom MA, Notredame C -
Using Multiple Alignment Methods to Assess the Quality of Genomic Data Analysis
Notredame and Abergel, in Bioinformatics and Genomes: Current Perspectives. M. Andrade, Horizon Scientific Press: pp 30-50.