T-RMSD Tutorial A quick start to the T-RMSD protein structural classification
Description
The T-RMSD web server allows a fine-grained structural classification of proteins using the T-RMSD method (Tree based on Root Mean Square Deviation), see [1, 2].
Providing a set of proteins sequences, it produces a structural tree with support values for each cluster node, reminiscent to bootstrap values. These values, associated with the tree topology, allow a quantitative estimate of structural distances between proteins or group of proteins defined by the tree topology.
Data input
-
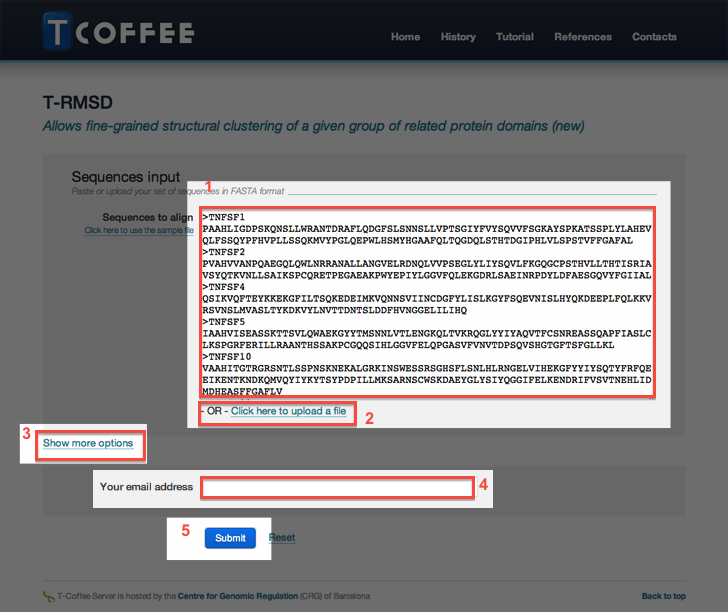
Enter the sequences in the input field (marked with '1' in the picture below) or alternatively
provide them using the Upload link (2).
The protein sequences have to be in a standard format, either FASTA or ClustalW. - You may access to T-RMSD advanced settings by clicking the link Show more options (3).
- Provide your email address (4) if you want to be notified when the job complete.
- Press the button submit (5) to submit your request.
Please note that the computation can take some minutes to be fulfilled. You don't need to keep your browser open to wait for the result. You can close it or navigate away to another page.
Use the History link, in top-bar menu to get access to the computation result at any moment.
Output interpretation
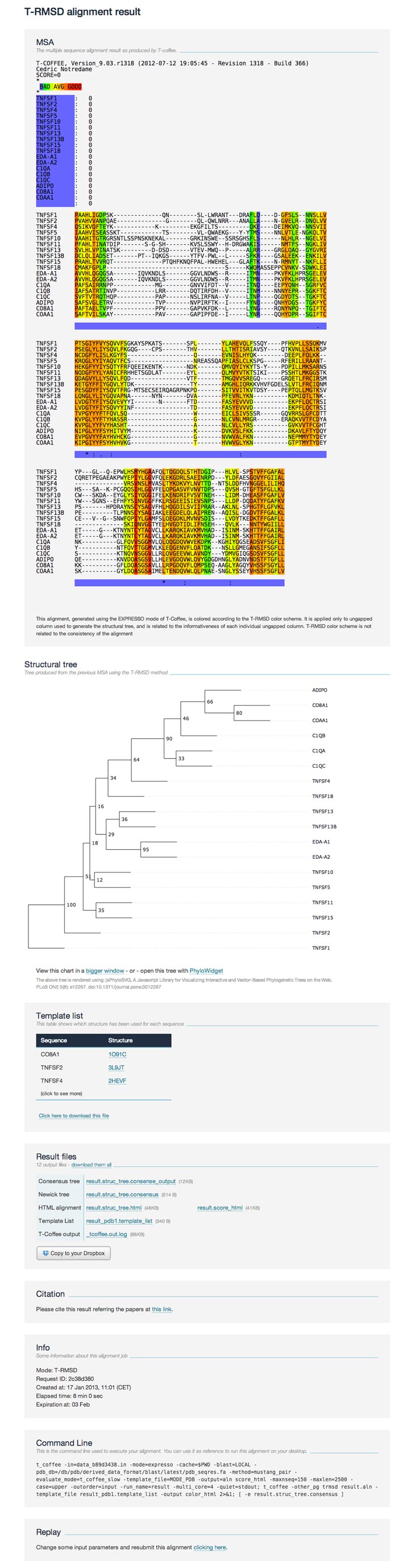
The result page contains the following sections:
- MSA the MSA, colored according to the T-RMSD scheme (it is not a T-Coffee consistency based color scheme). The T-RMSD color scheme is related to the informativeness of each ungapped column, from blue (poorly discriminant) to red (highly discriminant).
- Structural tree the resulting structural tree with its support values. It comes along with the option to modify the tree using an external application. The user can also download the Newick format tree file (available in the Result files section) to use other visualization tools
- Template list the list of template structure files associated with each sequence. The structure files are named after the PDB code and the identifier chain (ChainID) of the chain use for the computation. Structures are linked to their corresponding PDB webpage for more information.
- Result files the result files coming from the alignment and T-RMSD method; all files can be downloaded as a single zip file
- Citation the related article when citing the T-RMSD.
- Info some information related to the submitted job (running time, date, etc).
- Command line the command lines (separated by a semicolon) used by the webserver to run successively the Expresso alignment and the T-RMSD method. Lines can then be reused and/or modified to run locally with your own T-Coffee package.
- Replay this link allows the user to re-run the job while modifying input options or data.
References
- T-RMSD: a fine-grained, structure-based classification method and its application to the functional characterization of TNF receptors Magis C, Stricher F, van der Sloot AM, Serrano L, Notredame C.
- An improved understanding of TNFL/TNFR interactions using structure-based classifications. Magis C, van der Sloot AM, Serrano L, Notredame C.